Segvis
R
genomics
visualization
Segvis is a package to visualize genomic data around genomic regions.
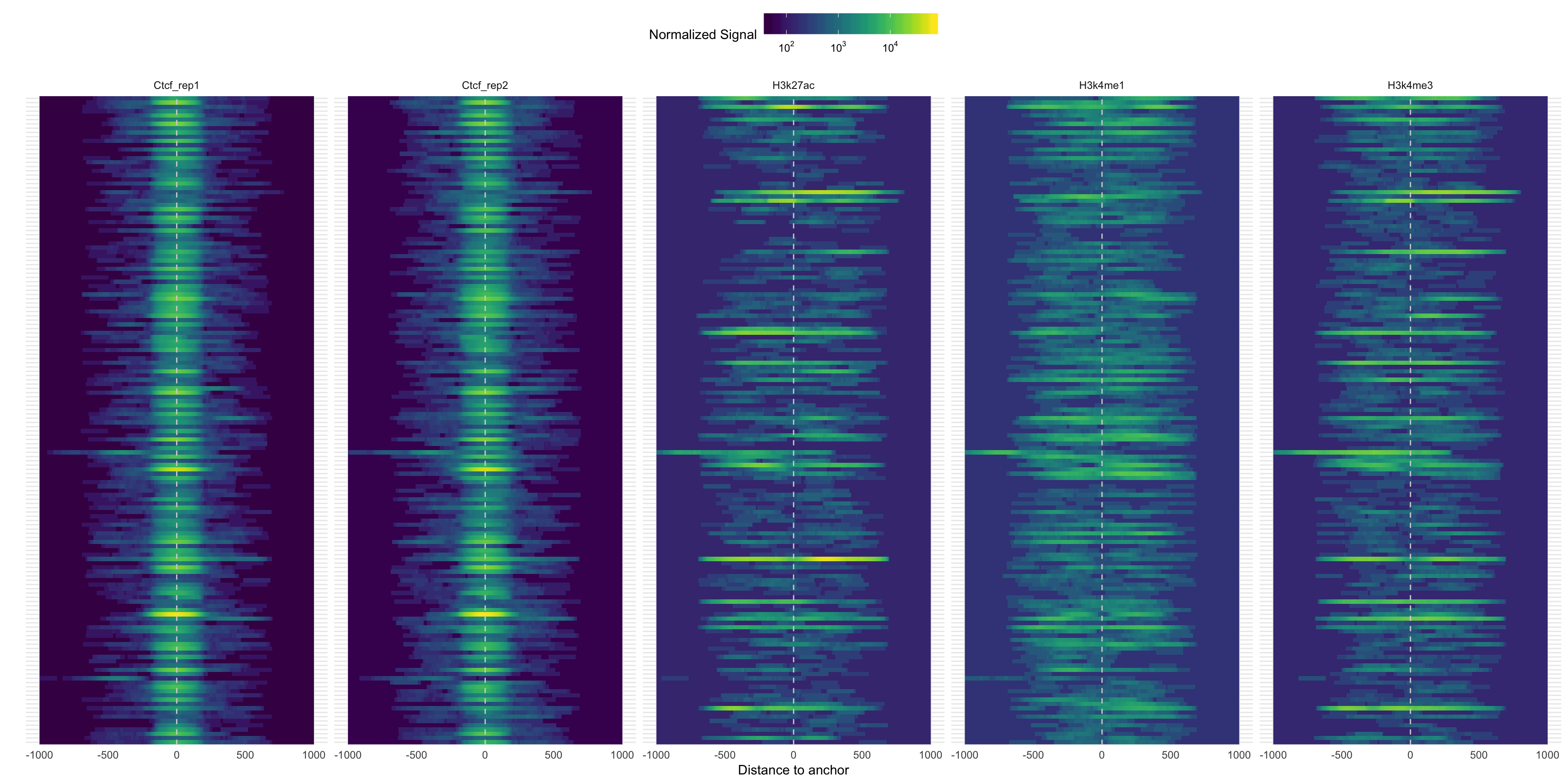
An R package to visualize ChIP-seq experiments across a set of pre-defined peaks. The package requires a set of pre-defined genomic regions, and aligned reads in bam format. We can do many tasks with it, some examples are:
- Extracts read data of specified input regions
- Plot peaks of different conditions across the same set of regions
- Plot central measures for a set of regions with the same width
- Subset regions according to used defined annotations
- Plot the heatmap of signal curves accross regions separated by annotation
The package is available here
The image below was generated with Segvis, and it shows the CTCF and histone experiments signal around the CTCF peaks’ summits.